This is what an LSTM unit approximately looks like:

I say approximately because it seems to vary depending on the implementation whether the input goes through a non-linearity before getting to the gates, whether input is given to every gate, etc.
That image is from this page on Eric Yuan’s blog which has a great explanation of LSTMs, as does this page of Chris Olah’s blog.
I’m going to assume you’ve read those or something like them, and talk about the biological plausibility of LSTMs. This post began as a response to discussion in class over Thomas George’s question on this lecture page.
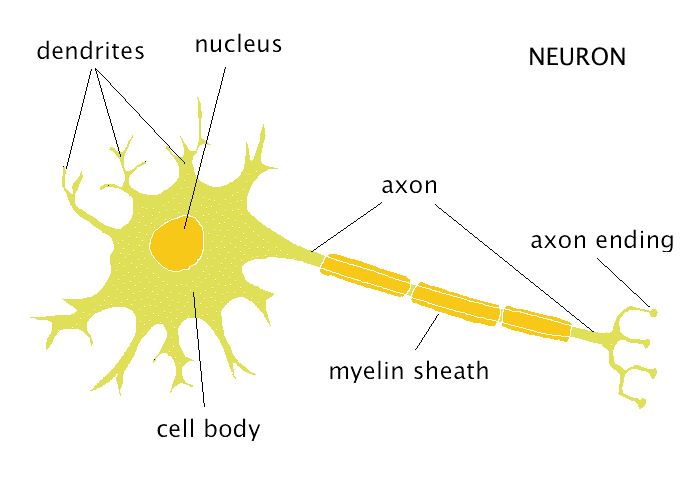
Just as a refresher, this is what a cartoon neuron looks like; generally we consider that the dendrites perform a weighted sum, the cell body performs some non-linear transformation of that sum, and the output is passed along the axon to other neurons (‘axon ending’ in this diagram generally ‘ends’ at a synapse with the dendrites of other neurons). Image from here.
Gating in general:
I think gating is like having some connections higher in the dendritic arbor (closer to the cell body), which have a proportionally very large impact on the processing done by the cell. So you unfold each gate into a sigmoid MLP with one output to the central cell, and then an LSTM is just a specific way to nest/restrict connectivity between neurons where the neurons can have different activation functions and different degrees of effect on each other.
In this view, the different LSTM variants (peepholes, coupled gates, etc.) are just different connectivity structures – in particular, peepholes allow connections between the gate-cells, and coupled gates use the same MLP for various gates.
To look at the three gates (“neuron types”) individially:
Input gate:
This is like the central cell having input from a neuron which has had inputs from the “real” input neurons x(t), and also from the “output” of the previous state h(t-1).
For example, this could be the case of a cell in the LGN (lateral geniculate nucleus) which receives information about light levels from sensory neurons in the retina, and also feedback information from the primary visual cortex.
Forget gate:
This is like the central cell having input from a neuron which looks at the same stuff the input gate does, and also has input from the central cell.
In class we talked about a differential formula for how voltage changes across the membrane:
where Vi is like an input to one’s self where the value depends on both the input and on the previous value (if you were to discretize time). In other words, makes the cell inertial to changing its value. I see this like giving a bias to the ‘central’ cell in this unfolded LSTM.
The above formula should actually be
where is a time constant. We talked in class about this time constant not being fixed in LSTMs, and whether or not this is biologically plausible. There are cells in the hippocampus whose spiking rates appear to modulate our perception of time, and these correlations are not fixed.
To continue the previous example, we can imagine that the forget gate is a cell in the LGN connected to the input gate cell I describe above, and which also has connections to the hippocampus and to the central cell. This paper talks about hippocampal connections to the LGN.
Output gate:
This is like the central cell having input from a neuron which looks at the same stuff as the input gate does, and attaches to the central cell not in the dendritic arbor but on the axon.
This paper talks about a region of hippocampal cells in particular having this structure (well, they observe that many axons in this area derive from dendrites, which amounts to the same thing).